Rosalind
- Problem solving learning platform
- Inspired by Project Euler e Google Code Jam
- Name commemorates Rosalind Franklin
- https://rosalind.info
Choosing problems
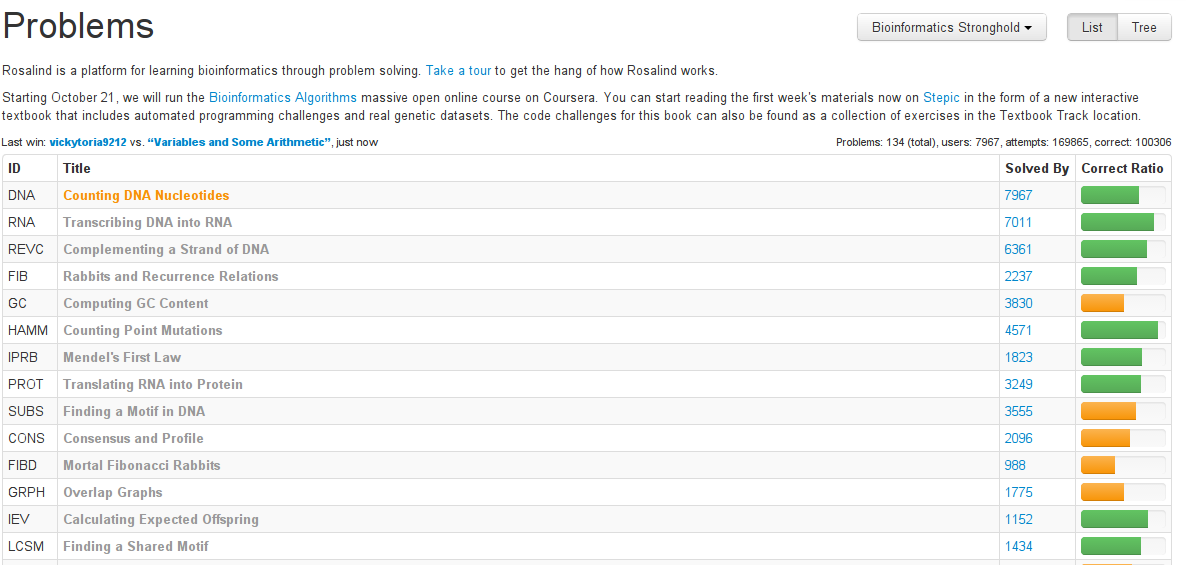
List
Choosing problems
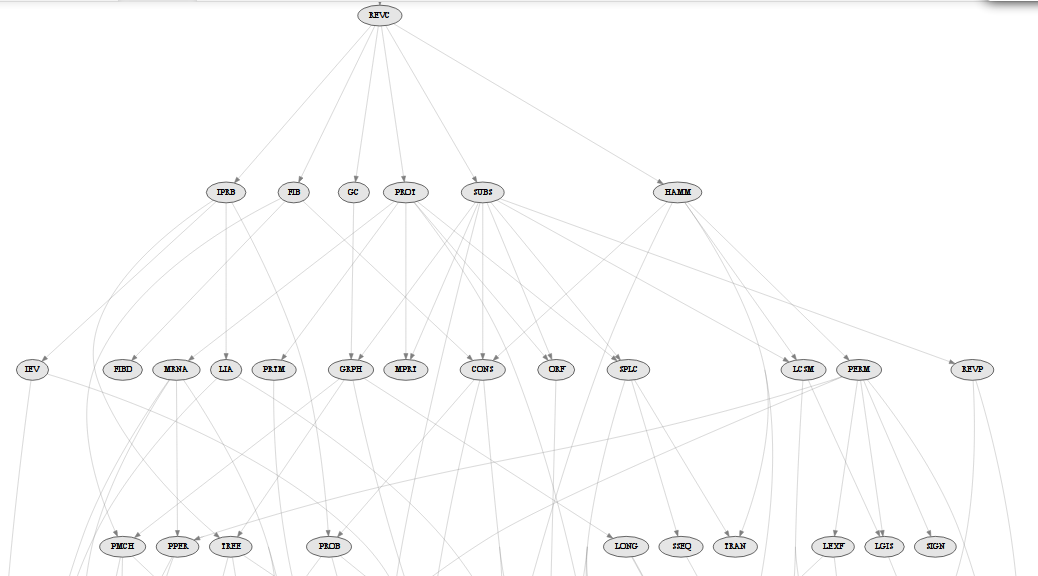
Tree
Choosing problems
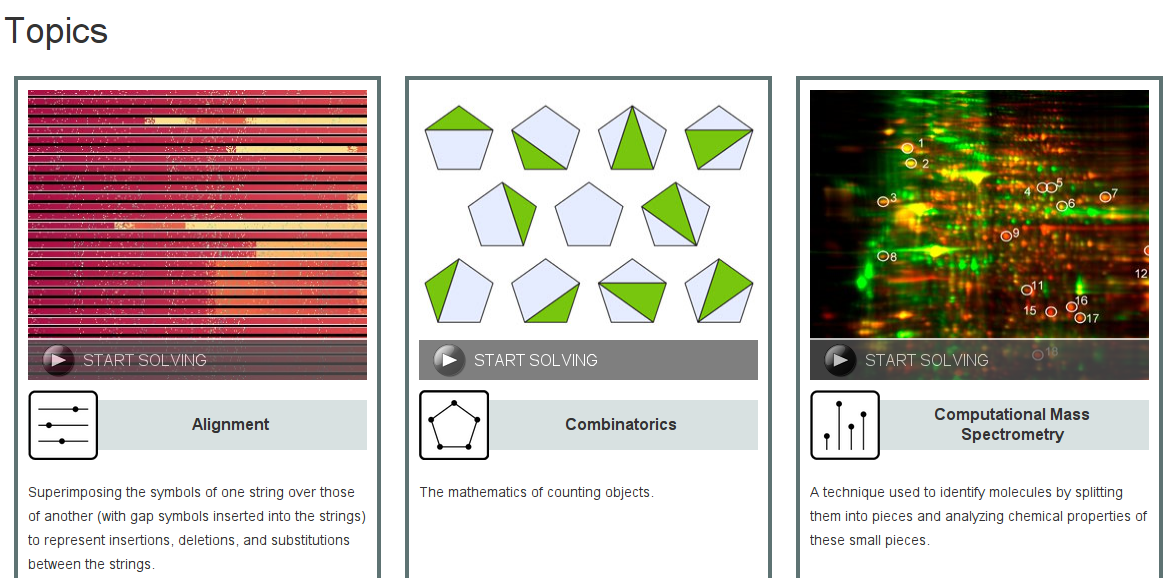
Topics
Choosing problems

Locations
Gamification!
Anatomy of a problem
- http://rosalind.info/problems/dna/
- Description
- Input
- Output
- Sample input
- Sample output
Anatomy of a problem
Submitting
Don't ignore the explanation!
Learning!
- Great for learning new languages
- Python 2/3
from __future__ import print_function, division
- Dicts differences (.items(), .values() and .keys() returns views in Python 3)
- from functools import reduce
- List comprehensions vs map/filter
List comprehensions provide an alternative to using the built-in map() and filter() functions. map(f, S) is equivalent to [f(x) for x in S] while filter(P, S) is equivalent to [x for x in S if P(x)]. One would think that list comprehensions have little to recommend themselves over the seemingly more compact map() and filter() notations. However, the picture changes if one looks at a more realistic example. Suppose we want to add 1 to the elements of a list, producing a new list. The list comprehension solution is [x+1 for x in S]. The solution using map() is map(lambda x: x+1, S). The part “lambda x: x+1” is Python’s notation for an anonymous function defined in-line.
It has been argued that the real problem here is that Python’s lambda notation is too verbose, and that a more concise notation for anonymous functions would make map() more attractive. Personally, I disagree—I find the list comprehension notation much easier to read than the functional notation, especially as the complexity of the expression to be mapped increases. In addition, the list comprehension executes much faster than the solution using map and lambda. This is because calling a lambda function creates a new stack frame while the expression in the list comprehension is evaluated without creating a new stack frame.
http://python-history.blogspot.com.br/2010/06/from-list-comprehensions-to-generator.html
- return map(dna.count, order)
+ return [dna.count(o) for o in order]-from itertools import starmap
-import operator
def hamming(s1, s2):
- return sum(starmap(operator.ne, zip(s1, s2)))
+ return sum(a != b for a, b in zip(s1, s2))- Context manager for file handling
with open('rosalind_dna.txt') as dataset:
dna = dataset.readline().rstrip()
- Sets FTW!
with open(os.path.join('data', 'rosalind_dbru.txt')) as dataset:
dna_set = {s.rstrip() for s in dataset}
reverse_complements = {reverse_complement(s) for s in dna_set}
dna_strings = dna_set | reverse_complementsRelated
- https://www.coursera.org/course/bioinformatics
- Started in October, already ending =/
- Online textbook available:
https://stepic.org/Bioinformatics-Algorithms-2/
Questions?
luizirber.org
luiz.irber@gmail.com