Por que Python é lento?
- Tipagem dinâmica
- Lookup de atributos
- a[1]
- Mesmo no Numpy
Qual a abordagem atual?
- Escrever parte crítica em C/C++/Fortran e encapsular
- SWIG
- ctypes
- Cython
- f2py
- CPython API
- Escrever diretamente em Cython
- Depois de aprender os atalhos e detalhes...
Mas... não dá para ser mais fácil?
Objetivos do Numba
- Funcionar com CPython (e Numpy, Scipy e todo o stack científico)
- Modificações mínimas no código (inferência de tipos)
- O programador decidir o que deve ser acelerado ou não
- Possibilitar a criação de extensões estáticas (para bibliotecas)
- Produzir código tão rápido quanto C (até mesmo Fortran?)
- Suportar array-expressions do Numpy e criação de ufuncs
- Produzir código para hardware vetorial (GPUs, aceleradoras, many-core)
LLVM
- Provê a infraestrutura
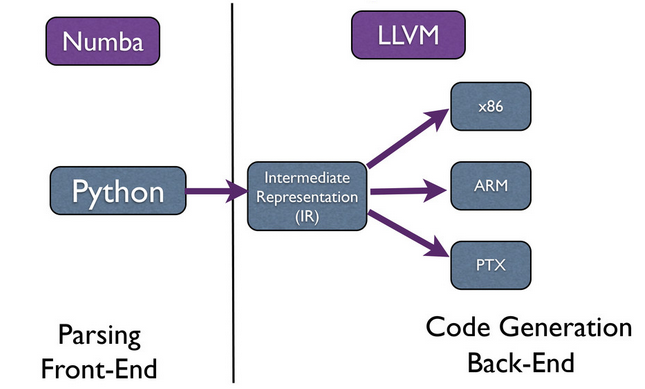
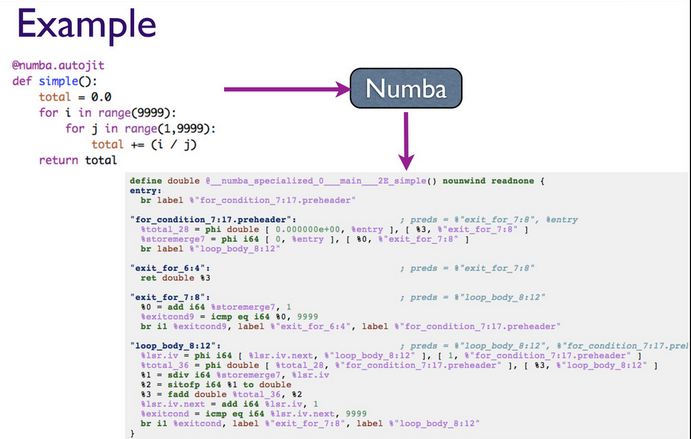
API simples
- Dois decoradores:
- jit: programador especifica tipos
- autojit: detecta tipos de entrada, saída, gera código se necessário, e executa
from numba import autojit, jit, double
#@jit('void(double[:,:], double, double)')
@autojit
def numba_update(u, dx2, dy2):
nx, ny = u.shape
for i in xrange(1,nx-1):
for j in xrange(1, ny-1):
u[i,j] = ((u[i+1,j] + u[i-1,j]) * dy2 +
(u[i,j+1] + u[i,j-1]) * dx2) / (2*(dx2+dy2))
Comparando desempenho
http://jakevdp.github.io/blog/2013/06/15/numba-vs-cython-take-2/
"As before, I'll use a pairwise distance function. This will take an array representing M points in N dimensions, and return the M x M matrix of pairwise distances. This is a nice test function for a few reasons. First of all, it's a very clean and well-defined test. Second of all, it illustrates the kind of array-based operation that is common in statistics, datamining, and machine learning. Third, it is a function that results in large memory consumption if the standard numpy broadcasting approach is used (it requires a temporary array containing M * M * N elements), making it a good candidate for an alternate approach."
import numpy as np
X = np.random.random((1000, 3))
Python puro
def pairwise_python(X):
M = X.shape[0]
N = X.shape[1]
D = np.empty((M, M), dtype=np.float)
for i in range(M):
for j in range(M):
d = 0.0
for k in range(N):
tmp = X[i, k] - X[j, k]
d += tmp * tmp
D[i, j] = np.sqrt(d)
return D
Numpy com broadcast
def pairwise_numpy(X):
return np.sqrt(((X[:, None, :] - X) ** 2).sum(-1))
Numba
from numba.decorators import autojit
pairwise_numba = autojit(pairwise_python)
@autojit
def pairwise_numba(X):
...Cython otimizado
%load_ext cythonmagic
%%cython
import numpy as np
cimport cython
from libc.math cimport sqrt
@cython.boundscheck(False)
@cython.wraparound(False)
def pairwise_cython(double[:, ::1] X):
cdef int M = X.shape[0]
cdef int N = X.shape[1]
cdef double tmp, d
cdef double[:, ::1] D = np.empty((M, M), dtype=np.float64)
for i in range(M):
for j in range(M):
d = 0.0
for k in range(N):
tmp = X[i, k] - X[j, k]
d += tmp * tmp
D[i, j] = sqrt(d)
return np.asarray(D)
Fortran (não otimizado!)
%%file pairwise_fort.f
subroutine pairwise_fort(X,D,m,n)
integer :: n,m
double precision, intent(in) :: X(m,n)
double precision, intent(out) :: D(m,m)
integer :: i,j,k
double precision :: r
do i = 1,m
do j = 1,m
r = 0
do k = 1,n
r = r + (X(i,k) - X(j,k)) * (X(i,k) - X(j,k))
end do
D(i,j) = sqrt(r)
end do
end do
end subroutine pairwise_fort
Overwriting pairwise_fort.f
# Compile the Fortran with f2py.
# We'll direct the output into /dev/null so it doesn't fill the screen
!f2py -c pairwise_fort.f -m pairwise_fort > /dev/null
from pairwise_fort import pairwise_fort
XF = np.asarray(X, order='F')
Scipy
from scipy.spatial.distance import cdist
Scikit-learn
from sklearn.metrics import euclidean_distances
Medindo tudo!
%%capture timeit_measures
%timeit pairwise_python(X)
%timeit pairwise_numpy(X)
%timeit euclidean_distances(X, X)
%timeit cdist(X, X)
%timeit pairwise_cython(X)
%timeit pairwise_fort(XF)
%timeit pairwise_numba(X)
timeit_measures.show()
1 loops, best of 3: 7.19 s per loop 10 loops, best of 3: 41.7 ms per loop 100 loops, best of 3: 16.4 ms per loop 100 loops, best of 3: 7.77 ms per loop 100 loops, best of 3: 7.36 ms per loop 100 loops, best of 3: 7.77 ms per loop 1 loops, best of 3: 6.86 ms per loop
Massageando os dados
from pint import UnitRegistry
ureg = UnitRegistry()
labels = ['python\nloop', 'numpy\nbroadc.', 'sklearn', 'scipy', 'cython', 'fortran/\nf2py', 'numba']
times = []
for line in timeit_measures.stdout.split('\n')[:-1]:
value, unit = line.split(':')[-1].split('per')[0].strip().split()
times.append(ureg.Quantity(float(value), unit).to('seconds'))
measures = list(reversed(sorted(zip(times, labels))))
measures
[(<Quantity(7.19, 'second')>, 'python\nloop'), (<Quantity(0.0417, 'second')>, 'numpy\nbroadc.'), (<Quantity(0.0164, 'second')>, 'sklearn'), (<Quantity(0.00777, 'second')>, 'scipy'), (<Quantity(0.00777, 'second')>, 'fortran/\nf2py'), (<Quantity(0.00736, 'second')>, 'cython'), (<Quantity(0.00686, 'second')>, 'numba')]
Comparando resultados
%pylab inline
Populating the interactive namespace from numpy and matplotlib
WARNING: pylab import has clobbered these variables: ['double'] `%pylab --no-import-all` prevents importing * from pylab and numpy
labels = [m[1] for m in measures]
timings = [m[0].magnitude for m in measures]
x = np.arange(len(labels))
ax = plt.axes(xticks=x, yscale='log')
ax.bar(x - 0.3, timings, width=0.6, alpha=0.4, bottom=1E-6)
ax.grid()
ax.set_xlim(-0.5, len(labels) - 0.5)
ax.set_ylim(1E-3, 1E2)
ax.xaxis.set_major_formatter(plt.FuncFormatter(lambda i, loc: labels[int(i)]))
ax.set_ylabel('time (s)')
ax.set_title("Pairwise Distance Timings")
<matplotlib.text.Text at 0x42b6950>
Apresentação executável
https://github.com/luizirber/pythonbrasil9/blob/master/numba.ipynb
- Anaconda para dependências
- pip install pint
Obrigado
luizirber.org
luiz.irber@gmail.com